DNA replication in eukaryotes
Defining the eukaryotic replisome in live cells
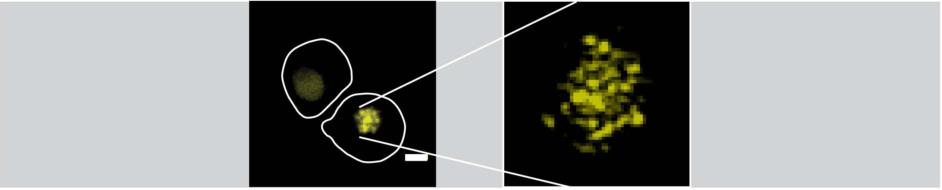
Outline of two S. cerevisiae cells labelled with PCNA-mNeonGreen (left), one of them undergoing S-phase. A close-up view of a Z-projection of the replicating nucleus obtained by Structural Illumination (right). Multiple fluorescent foci representing each one or more replisomes are observed.
It has been only in the last decade that we have finish identifying (probably) all the subunits of the replisome in yeast. Great efforts in the community are now focused in understanding how these components assemble into a functional machine. Our lab aims at contributing to understand the architecture of the replisome, the inner dynamics in the replisome, and how its coordinated function results in the synthesis of the chromosome. Achieving a high signal-to-noise ratio in fluorescence microscopy for quantitative approaches is more complicated than in bacteria due to higher levels of endogenous and out-of-focus fluorescence in yeast. Our lab is developing new microscopy, genetic and labelling approaches that allows the study of live yeast using single-molecule and super-resolution microscopy techniques.
In addition to the understanding of the replisome, we are also working on the mechanisms that regulate origin firing during S-phase. S. cerevisiae has hundreds of potential origins, many of which fire at different stages of S-phase. Origin selection is established by multiple factors that include DNA sequence, chromatin factors and the action of limiting initiation factors. We are currently working to determine the distribution in the genome and the kinetics of initiation factors.
We have characterised the movement of nuclei (green) inside growing budding yeast to helps us plan experiments where we follow single-molecules bound to DNA. In the movie above the vacuole (red) was also labeled. A picture was taken every second.
