Current Projects
Replisome architecture, dynamics, and mutation avoidance
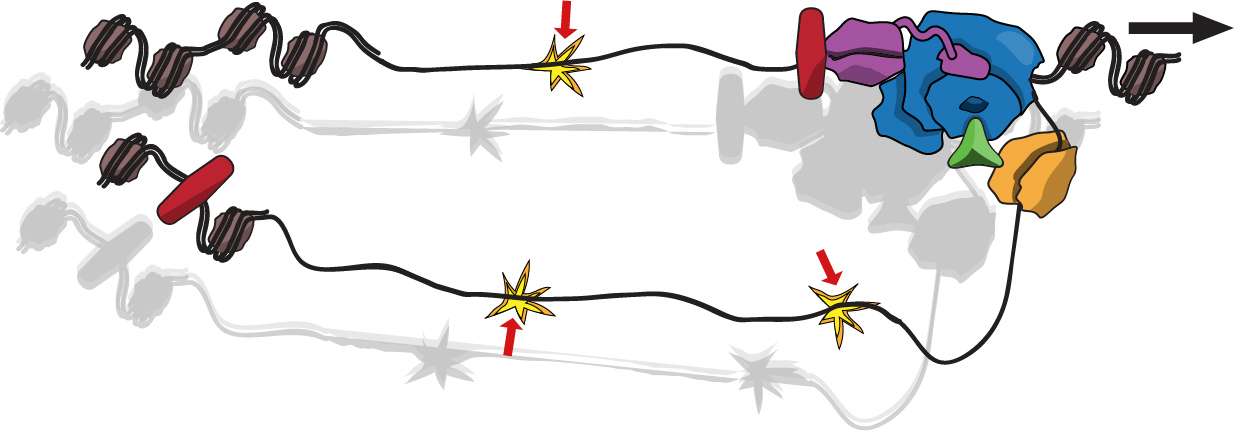
The most crucial function of the replisome is to coordinate the activities of DNA unwinding and DNA synthesis. Failure can result in the excessive accumulation of single strand DNA, which is a source of mutation and is more prone to breaks. We hypothesize that the architecture and dynamics in the replisome play a role in the coordination of DNA unwinding and DNA synthesis. Hence, current work in our group aims to understand these aspects of the replisome and their link to genome integrity in both, bacteria and budding yeast.
We use single-molecule microscopy to characterize the replisome in live cells. We also generate custom scripts and apply machine learning approaches to analyse the images acquired. These approaches have led us to describe the stoichiometry of the active replisome and to the surprising demonstration of a fast turnover (with a timescale of few seconds) of the DNA polymerase in bacteria. We are currently working to understand what triggers the unbinding of the DNA polymerase from DNA, the mechanisms that facilitate its recruitment back to DNA, and the benefits of this fast turnover to the cell.
We also established single-molecule live cell approaches in yeast to show that the DNA polymerases are much more tightly bound to the rest of the replisome (turning over in a timescale of few minutes). Current work aims to determine some of the protein-protein interactions holding the replisome together and to extend the characterization to other proteins associated to the replisome.
- Kapadia et al, 2021, Nucleic Acids Res 49:e79
- Kapadia et al, 2020, Mol Cell 80: 114
- Soubry et al, 2019, PNAS 116: 11747
- Beattie and Kapadia et al, 2017, eLife 6 DOI: 10.7554/eLife.21763
- Reyes-Lamothe et al, 2010, Science 328:498
Chromosomes, DNA replication and the cell cycle
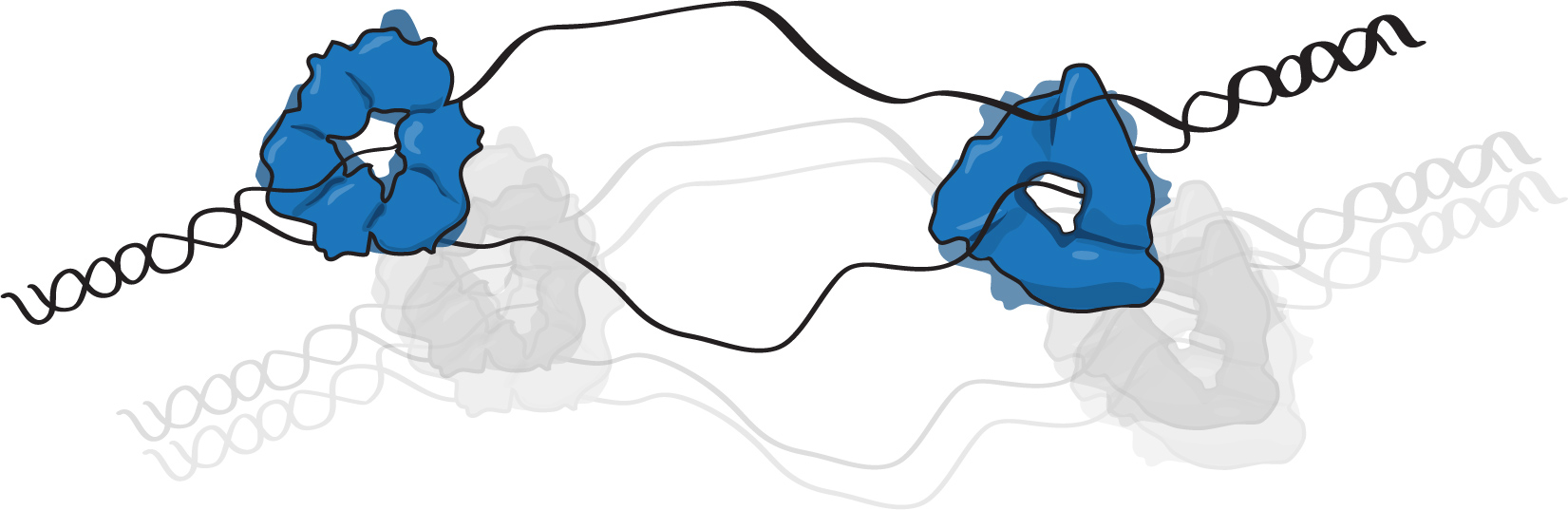
Our lab is interested in understanding how cell regulate the assembly and activation of the replisome, which is a crucial aspect to maintain the integrity of the genome. In bacteria, we are studying the activity and regulation of the initiation protein, DnaA. While in budding yeast we are working on the characterization of factors that control the timing of firing at different origins.
- Reyes-Lamothe and Sherratt, 2019, Nature Reviews Microbiol 17: 467
Cell size control
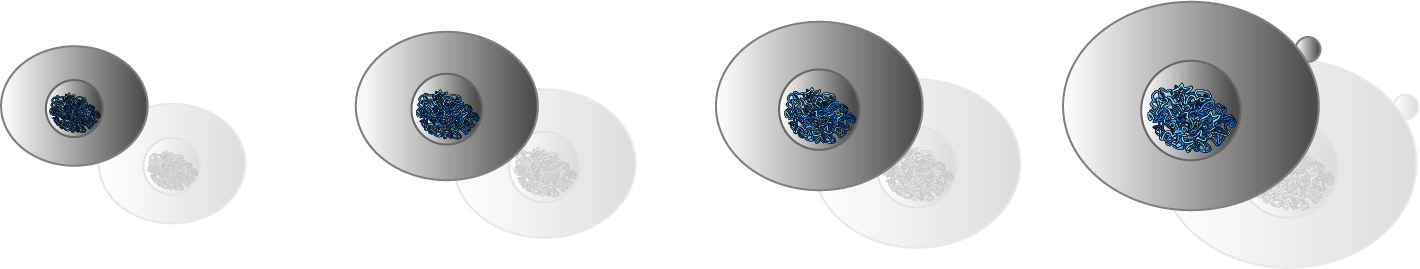
We also collaborate with the group of Jan Skotheim (Stanford University) to study how cells sense and respond to cell size in budding yeast. Currently we focus on the regulation of the transcription factor SBF by the inhibitor protein Whi5, which establish the time of the cell cycle ‘Start’ in G1.
- Swaffer et al, 2021, Cell 186: 5254
The cell biology of antibiotic resistance
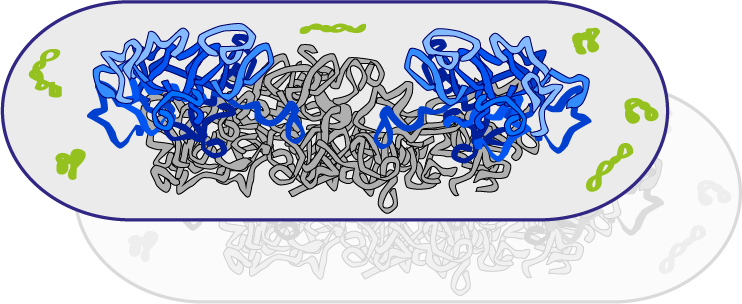
In collaboration with the group of Marcelo Tolmasky at CSU Fullerton, we are studying how antibiotic resistance is established in the bacterial cell. In recent published work we determined the distribution of a multi-resistance plasmid in cells, showing that plasmids preferentially localize at the edges of the cell, where there is a low density of chromosomal DNA. We are currently using fluorescence microscopy to measure the activity of the antibiotic resistance enzymes in live cells. We aim to establish enzymatic kinetics in vivo and determine the cellular factors that influences it.
- d'Udekem d'Acoz et al, 2024, mSphere: e0078923
- Reyes-Lamothe et al, 2014, Nucleic Acids Res 42:1042
