DNA Pol Evolution
Directed evolution of DNA Polymerase
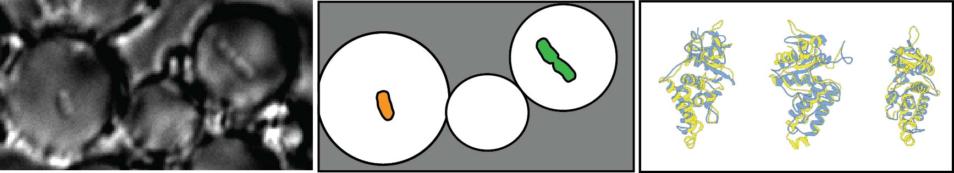
Cells expressing mutant forms of DNA Pol III are trapped in bubbles of water-in-oil emulsion containing PCR mix (left). Outline of two bubbles containing a single cell each (middle). Three different views of the palm domain's structure of Pol III (right).
Our lab constantly searches for new tools to stimulate controlled responses from cells that can help us understand their biochemistry. An important point of control in our studies is the replicative polymerase, DNA Pol III, which is in charge of synthesizing the chromosome. Pol III has evolved to have very high fidelity at incorporating nucleotide residues during the elongation of DNA. In good measure, this is due to high efficiency at discriminating between substrate nucleotides. However, in the lab this substrate specificity limits our capacity to track the Pol III activity in cells. Bulky base analogues, like those coupled to a fluorescent dye, will not be incorporated into DNA, even when the base they carry is one of the four natural bases for DNA. In order to obtain new ways to track and manipulate Pol III activity, we are using directed evolution techniques to generate Pol III mutants that maintain high fidelity of incorporation but relaxed or different nucleotide selectivity
